Screen of PlGF Peptide for Targeted Imaging of Ovarian Cancer Cells
Li Yan1$, Rongrong Zhang3$, Junwei Zhao1, Jiachang Hu1, Lingfei Han1, Sumei Niu1, Qiuqie Ba1, Yi Guo1, Fang Fang1, Fanfei Kong1, Jing Sun1, Chao Lin2, Fang Li 1*
1Department of Gynaecology, Shanghai First Maternity and Infant Hospital, Tongji University School of Medicine, No.536, Chang Le Road, Jing An District, Shanghai, 200040,People’s Republic of China.
2Institute for Biomedical Engineering and Nano Science, Tongji University School of Medicine, Shanghai 200092, China.
3Department of Gynaecology,Xin Hua Hospital,Jiao Tong University School of Medicine.
$These authors contribute equally to the article.
Abbreviations
PlGF Placental growth factor
DMEM Dulbeccl’s Modified Eagle’s Medium
Amp Ampicillin
Ab Antibody
Ag Antigen
bp Base pair
BSA Bovine serum albumin
cDNA Complementary DNA
DEPC Diethyl pyrocarbonate
EDTA Ethylene diaminetetraacetic acid
IPTG Isopropylthio-β-D-galactoside
Kan Kanamycin
LB Luria-Bertani medium
MCS Multiple cloning sites
PBS Phosphate-buffered saline
PCR Polymerase chain reaction
RBS Ribosome binding site
TBE Tris-borate EDTA buffer
VEGF Vascular endothelial growth factor
VEGFR Vascular endothelial growth factor receptor
X-gal 5-bromo-4-chloro-3-indolyl-β-D-galactoside
Summary
Objective: This study screened a human placenta growth factor (placental growth factor, PlGF)-specific binding peptide, which is the theoretical basis for further study of the function of PlGF and its role in the targeted therapy and early diagnosis of ovarian cancer.
Methods: Human placenta growth factor (placental growth factor, PlGF) was selected as the target antigens for the PCR amplification to get a DNA library containing random 10-peptide. Ribosome display technology was used to screen the DNA library. The affinity between the 10-peptide and target proteins was confirmed by ELISA method, and its affinity constant was calculated. FITC-labeled and Quantum dot-labeled 10-peptide was tested by flow cytometry and confocal microscope respectively to confirm the targeted binding capacity in vitro with the ovarian cancer cell line (of SKOV-3).
Results: The DNA library we constructed has a storage capacity of 1.8 × 1012. We get five PlGF-specific binding peptide, named respectively P1, P2, P3, P4 and P5. Their basic sequence were TL / FS〠WPA / TRSTS〠WNWP and TS. P5 was confirmed to has a high binding activity with the target antigen PlGF. The functional affinity constant is (3.000 ± 0.253) x 106M-1. There is a strong binding capacity of up to 94.19% between P5 and ovarian cancer cells. P5-peptide conjugated quantum dot could recognize SKOV-3 cells specifically.
Conclusion: PlGF protein binding peptides collection is available through ribosome display technology. The peptides has a strong binding activity with PlGF which could be used for the targeted therapy of ovarian cancer and early diagnosis.
Introduction
Ovarian cancer is one of common female reproductive system tumors. It is characterized by high incidence, high degree of malignancy, high mortality, infiltrative growth of tumor, rapid metastasis, poor response to drug treatments[1]. About 70% of patients were diagnosed at advanced stages. In order to promote the early diagnosis and treatment of ovarian cancer, ovarian cancer-specific markers and therapeutic targets related research is currently one of the hotspots[2].
PlGF is an important member of the VEGF family. It promotes angiogenesis through regulation of endothelial cells, smooth muscle cells and hematopoietic stem cells [3]. A number of studies confirmed that PlGF expression increased in a variety of tumor tissues, and it’s related with tumor stage, metastasis and survival rate [4]. Some studies have confirmed that the targeted PlGF or its receptor (VEGFR-1) immune therapy can inhibit tumor growth [5-7], indicating that PlGF is feasible as a cancer therapeutic target. The present immunotherapy drugs are mostly monoclonal antibody-based, which are easily inducing immune responses in the human body. They may be remained in the blood circulation instead of effective tumor tissue enrichment, because of their poor tissue penetration [8], thus losing the role of the “oriented” treatment. The development and application of the polypeptides oriented carrier may provide new approach to address this critical issue and therefore, screening of specific target gene binding peptide has become a research hotspot. In our preliminary experiments, we constructed single chain antibody library of ovarian cancer; screening with ribosome display technology, successfully got anti-PlGF single-chain antibody genes [9]. In the present study, we furtherly use the ribosome display technique to screen PlGF specific binding polypeptide, and to confirm the functional affinity constant, and thus provide a new method which has a broad prospect for the research and application of small molecule therapeutic agents.
Materials and methods
Materials
Cloning vector (pUCmT-vector) and escherichia coli strain (DH5a) are offered by Shangon Biotech (Shanghai) co., Ltd; DNA gel recycling kit and plasmid extraction kit are provided by Axygen Biotechnology Limited; In vitro rapid transcription kit/ Translation system kit (TNT T7 Quick for PCR DNA) are from Promega; Tosyl-activated DynabeadsM-280 is bought from Dynal Biotech ASA (Norway); PLGF is from Borcon Biotech Co.,Ltd; First strand cDNA Synthesis Kit: from Fermentas. Peptides are synthesized by GL Biochem (Shanghai) Ltd; Unless specified, chemicals were purchased from Sigma- Aldrich (St. Louis, MO) and used without further purification; Ovarian cancer cell lines (SKOV3) were bought from Shanghai Institute for Biological Science (CAS), and conserved in our laboratory. McCOY’S-5A medium, trypsin, fetal calf serum, penicillin and streptomycin are from Gibco (US).
Construction of DNA templates
The random oligonucleotide template was synthesized as a single-stranded 93-mer with the following sequence: 5’-ATA AAG CTT GCG AAA CAC (NNM)10 AAG GGA TCC GAC TAC AAA GAT GAC GAT GAC CAT TGG TCC ATG GGT -3’, where the central (NNM)10 represents random oligonucleotides. M may be G or T, N may be A, T, G or C. The forward primer of the first round of PCR: U15’-G CTC GTA TAA TGT GTG GCC ACA ACG GAA GGA GAT ATG ATA AAG CTT GCG-3’(49bp); The forward primer of the second round of PCR: U25’-GCG CTG CAG TAA TAC GAC TCA CTA TAG GGA GAG CTC GAT TAA TGT GTG-3’(48bp); The reverse primer: D15’-G GCA TAC ACT TGA AGG TGA GCC ACT ACC ACT TCC ACT ACC CAT GGA CCA ATG-3’(52bp). After two rounds of PCR under CR reaction condition (95℃, 5 min, 95℃30s,65℃ 30s, 72℃30s,30 circulation), and 10 min extension under 72℃, the products were detected by agarose gel electrophoresis, recycled by rubber tapping, and then sequenced. The products of two rounds PCR were directly transcribed and translated in vitro. The procedure is summarized as Figure 1.
Ribosome Screening
TNT T7 Quick for PCR DNA was used for in vitro transcription and translation. Protein antigens were directly connected with tosyl-activated Dynabeads. All procedures were in accordance with the manual operation under 30 ℃ bath heat, the mixture (40 μl TNT T7 Quick for PCR DNA, 2.5 μl random DNA library, 1 μl 1 mM methionine, 0.5 μl 100 mM magnesium acetate, add with DEPC to 50 μl ) was heated in the constant temperature water bath for 30min. 40U DNaseI and 6 μl 10× DNase digestion buffer were then added into the obtained mixture (400 mM Tris-HCl, pH=7.5, 60 mM MgCl2,100 mM NaCl), and ddH20 was added to its final volume of 60 μl. Incubated for 15min under 30 ℃, the above mixture was added the precooling 60 μl 2× dilution buffer (PBS containing 10 mM magnesium acetate) to be diluted.
Adding 1~2 μl prepared antigen-beads solution into the mixed solution, and jiggling it for 1 to 2 h under 4 ℃. Eluting the magnetic bead for five times with 10 μl elution buffer (PBS containing 5mM magnesium acetate and 0.1% Tween20). Eluting the magnetic bead for two times with 100 μl precooling no enzyme water, and adding 10 μl precooling no enzyme water heavy suspension magnetic beads for RT-PCR. The PCR products were used for the next circulation, referred to as transcription – translation – screening. The screening was conducted for five rounds (Figure 1).
Cloning and sequencing
After the selection, RT-PCR products were sub-cloned into pUCM-T, which was then transformed into E. coli DH5α. Selecting the clones due to the principle of α-complementary for sequencing to analyze the DNA sequences. Plasmid DNA was isolated, purified from E. coli DH5α, and subsequently analyzed by sequencing.
Synthetic peptide binding assays
The binding activities of peptides were determined using ELISA as described [9], with minor modifications. Wells were coated with PlGF,after binding of synthetic biotinylated peptides, horseradish peroxidase-linked streptavidin (HRP-streptavidin) was added, and binding was detected via the HRP enzymatic activity. Peptide-streptavidin conjugates were prepared by mixing HRP- conjugated streptavidin with peptides in PBS/0.1% BSA/0.2% Tween-2, then the mixture was incubated at room temperature for 30 min. The peptides were omitted in negative-control preparations.
Affinity Constant of P5
The affinity constant of P5 was determined by non-competitive enzyme immunoassay. ELISA plates were coated with 2, 1, 0.5 or 0.25 μg of PLGF. 100 μl of biotinylated P5 at concentrations of 15.625, 31.25, 62.5, 125, 250, 500 or 1000 ng/ml was added to the ELISA plate. Detection was performed with an HRP-streptavidin. Plates were developed with TMB-detection-solution and read at OD450 nm (Figure 3B). The concentration of P5 at OD50 according to the antigen-antibody reaction with different concentrations of PLGF was calculated. We got the affinity constant using the formula K= (n-1)/(nAb’-Ab), where Ab and Ab’ denote the concentration (mol/L) of peptide at OD50 when the concentrations of PLGF are Ag and Ag’, respectively.
Binding Characteristics in vitro study
Human ovarian cancer cell line SKOV3 cells were cultured (37 ℃, 5% CO2) in McCOY’S-5A medium, supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin. Cells were sub-divided and seeded into 24-well plastic culture dishes with 10,000 cells/ dish and incubate overnight. Cells cultured with McCOY’S-5A medium with 0.25% FBS for 4 h, then blocked with 1% BSA-McCOY’S-5A medium for 5 min. Cells were treated with 200 nmol/L FITC- conjugated P5 for 4 h, or 200 nmol/L P5 for 4 h followed by 200 nmol/L FITC- conjugated P5 for 4 h, respectively, the peptides were removed and washed with PBS for three times. Finally fluorescence image of cells was obtained with a Nikon inverted microscope (Nikon TE2000-S, Japan) and FACS.
Preparation of P5 peptide conjugated quantum dots
The PMAL-capped quantum dots (QDs) were prepared using a method reported in our previous work [10]. The prepared QDs were purified twice by ultracentrifuge at 45 000 rpm for 30 min. The synthesized QDs were dissolved in PBS for further experiments. P5 peptide was conjugated with QDs (at 605 nm wavelength) by using standard EDC-NHS reaction, at start molar ratio of PMAL-QDs to P5 peptide as 20:1 (P5: QDs),the conjugates were then purified by ultracentrifuge twice. The dynamic radii and zeta potential of the QDs were measured by using a nanoparticle size analyzer (Malvern 2000, Malvern, England). The electrophoretic mobility of the prepared nanoparticles was determined by 0.8% agarose gel in TBE buffer at 100 mV for 30 min. The morphology and core sizes of QDs and QD–P5 were studied by TEM (JEOL. Ltd Inc, Japan). The QDs and QD–P5 solutions were dropped onto a copper net and observed by TEM after dehydration.
Laser confocal microscopy image analysis
The SKOV3 and L99 fibroblast cells were seeded in a laser confocal microscopy 35 mm2 Petri dish (MatTek, USA) at a density of 1.0 × 105 cells and maintained for 48 h. QD-P5 complexes were prepared in serum-containing medium and incubated with cells for 4 h. Confocal images were obtained using Leica TCS SP5 confocal laser scanning microscope (Leica Inc., USA). To acquire fluorescence signal, each sample was excited at 450 nm with an Ar laser. A fluorescence signal of QD-P5 was detected at 488nm and 605 nm.
Statistics
At least three different biopsies were used per experiment and measurements were taken in triplicate. Data are presented as the mean and the standard error mean (SEM), and the statistical significance of differences were evaluated with the one-way anova text (* p < 0.05 and ** p<0.001).
Results
The designing and construction of DNA random library
The designed random oligonucleotides seen as the template, primers U1 and D went through the first round of PCR. Then with the product of first round of PCR as the template, primers U2 and D went through the second round of PCR (Figure 2A). The product of the second round of PCR was seen as the random DNA library for ribosome display, constructing and screening the random peptides library. After oligonucleotide going through two rounds of PCR, 167ã€199 bp-sized fragments were obtained (Figure 2B) . The number of recovered DNA fragments directly reflected the complexity of the produced libraries, which were up to 1.8×1012 members. In order to check the quality of the library, a small amount of a part of the 199-bp fragment was cloned, and 10 randomly chosen clones were sequenced. As expected, all the chosen clones were different (Figure 2C, Figure 2D).
The result of ribosome screening
A highly specific binding complex PRM( Protein ribosomal -mRNA) was obtained after five rounds of screenings. After RT-PCR and cloning, DNA was obtained and expanded. The monoclone was selected for sequencing to learn the characters of the highly specific binding peptides, and their common threads (Table1). We get five PLGF-specific binding peptide, named respectively P1, P2, P3, P4 and P5. Their basic sequence were TL / FS〠WPA / TRSTSã€WNWP and TS.
The affinity of peptides
Biotinylate the peptides for ELISA reaction with HRP marked streptavidin biotin, BSA was the negative-control preparations. The ratio of A450 value to 2.1 times higher than as a positive decision criteria. P5 could be seen to have a high affinity with the target antigen PLGF (Fig3A).
Figure 3B shows the dose-response curves of binding of the P5 to the PlGF. On the basis of the curves, the affinity constant of the P5 was calculated to be (3.000±0.253)×106M-1, according to the method described previously.
The Binding Characteristics in vitro study
The fluorescence images of live SKOV3 (human ovarian cancer cell line) after 4h incubation with 200 nmol/L FITC- conjugated P5 for 4 h , or 200 nmol/L P5 for 4h followed by 200 nmol/L FITC- conjugated P5 for 4h, respectively, shown in Figure 4A. A significant number of peptide was mainly distributed in cytoplasma.Flow cytometry analysis also support the same result, two groups of fluorescence intensity has statistically significant differences (Figure 4B,C). The binding between FITC-P5 and SKOV3 was sable, and could compete with unlabeled P5.
Characterization of QD-P5 conjugates and targeted imaging of ovarian cancer cells
Zeta sizer analysis showed that the particle size of QD-P5 was about 30 nm, and zeta potential was about -12 mV, switched the positive charge of P5 peptide after QD conjugation as showed in Figure 5. In order to confirm the targeted imaging of ovarian cancer cells, QD-P5 conjugates were incubated with SKOV3 cells and L99 cells respectively, then imaged by confocal microscope. Results showed that QD-P5 could specifically bind with SKOV3 cells while no binding with L99 fibroblast cells.
Discussion
Regulatory peptide analogues represent a class of molecules developed for specific cancer targeting. In spite of having a relatively short history of about 2 decades, they are now receiving increasing interest, as they are often advantageously compared with immune-targeting antibodies [11]. These peptides control and modulate the function of almost all key organs and metabolic processes, and include neuropeptides present in the brain, gut peptide hormones, as well as peptides present in vascular (vasoactive peptides) and endocrine systems [12]. Peptide’s receptors are proteins over-expressed in numerous human cancer cells. These receptors have been used as molecular targets for labeled peptides to localize tumour. Therefore, specific cancer targeting through selective peptides for diagnostic and therapeutic purposes is considered to be a promising strategy in oncology.
Angiogenesis is one of the crucial events for cancer development and growth[13]. Two members of the vascular endothelial growth factor (VEGF) family, VEGF-A and placental growth factor (PlGF), which are able to heterodimerize if coexpressed in the same cell, are both required for pathologic angiogenesis[14]. PlGF peptide could be a new candidate for antiangiogenic gene therapy in cancer treatment by inhibiting VEGF-dependent angiogenesis. Here, we screen one PlGF peptide candidate for ovarian cancer targeting by ribosome display technology. In order to confirm the targeted recognization to ovarian cancer cells, the PlGF peptide was conjugated with quantum dots, then conducted the targeted imaging of SKOV-3 cells by confocol microscope. Results showed the high affinity of PlGF peptide with ovarian cancer cells, and coud target SKOV-3 cells specifically. Quantum dots has been widely used in tumor imaging study due to their unique flurescent properties. Quantum dots served as an imaging reagent here for targeted recognization of ovarian cancer cells. For the targeted ovarian cancer therapy, biodegradable materials could be used to replace quantum dots as drug carrier, after conjugated with PlGF peptide.
Table 1. Sequences of peptide screened for PlGF ligands.
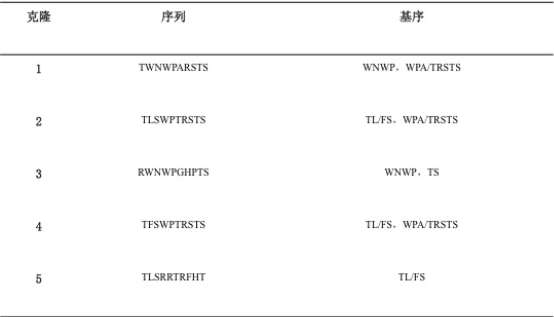
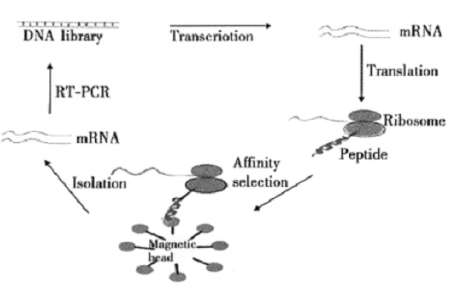
Figure 1. Schematic illustration of PlGF peptide screen.
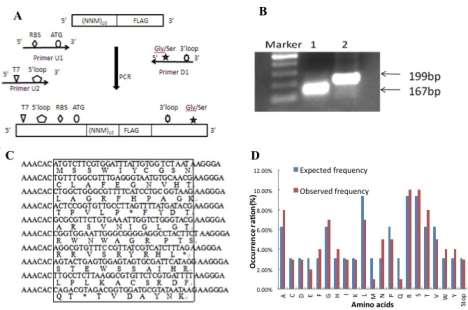
Figure 2. The designing and construction of DNA random library
A:
B:
C:
D:
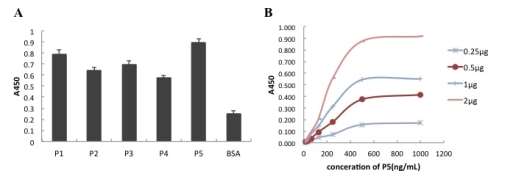
Figure 3. Affinity of P5 PlGF peptide with PlGF antigen and the dose-response curves of binding of the P5 to the PlGF.
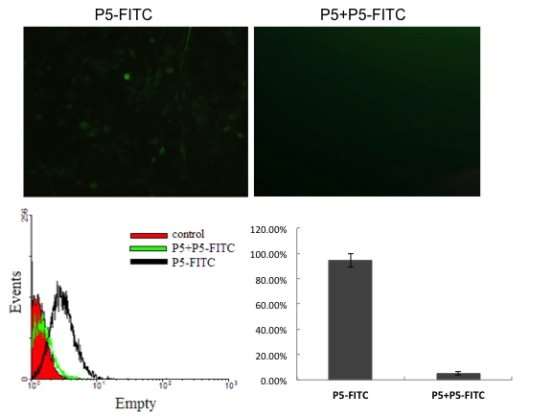
Figure 4. Fluorescence images of FITC labeled P5 peptide incubated with SKOV-3 cells, and followed flow cytometry analysis.
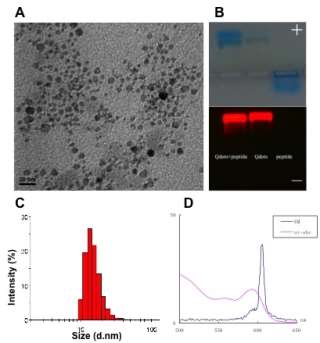 Figure 5. Characterization of P5 peptide conjugated peptide. A: TEM image; B: Agarose gel of P5 conjugated quantum dots, quantum dots only, and P5 peptide only from left to right side; C: Particle size of P5 conjugated quantum dots; D: Fluorescence and UV spectra of Quantum dots.
Figure 5. Characterization of P5 peptide conjugated peptide. A: TEM image; B: Agarose gel of P5 conjugated quantum dots, quantum dots only, and P5 peptide only from left to right side; C: Particle size of P5 conjugated quantum dots; D: Fluorescence and UV spectra of Quantum dots.
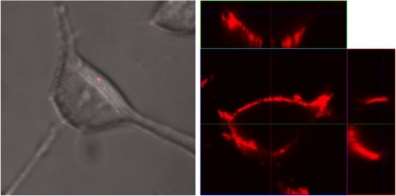
Figure 6. Confocal images of P5 peptide conjugated quantum dos incubated with L99 control cells (left) and targeted SKOV-3 cells respectively.
You have to be 100% sure of the quality of your product to give a money-back guarantee. This describes us perfectly. Make sure that this guarantee is totally transparent.
Read moreEach paper is composed from scratch, according to your instructions. It is then checked by our plagiarism-detection software. There is no gap where plagiarism could squeeze in.
Read moreThanks to our free revisions, there is no way for you to be unsatisfied. We will work on your paper until you are completely happy with the result.
Read moreYour email is safe, as we store it according to international data protection rules. Your bank details are secure, as we use only reliable payment systems.
Read moreBy sending us your money, you buy the service we provide. Check out our terms and conditions if you prefer business talks to be laid out in official language.
Read more