Analysis of Amino Acid Sequence and Identifying of Changes in the Influenza A/(H3N2) virus NS Gene in Isolates from Iran
Lawal D. Rogo, Farhad Rezaei, Nazanin Z. Shafiei Jandaghi, Nastaran Ghavami, Ghazal-Sadat Fateminasab, Talat Mokhtari-Azad
Subject: Medical Sciences
Keywords: Influenza A (H3N2) virus, Iran, Mutation, NS1 gene.
Abstract
The main functions of NS protein of influenza A virus is suppressing of type I IFN production by the host. Insight on the level of changes in the seasonal influenza A (H3N2) viruses NS gene in Iran is little. We sequence and analyzed amino acid of NS gene of 32 influenza A (H3N2) virus isolates for the purpose of getting detailed information about the genetic changes in this gene. Amino acid sequence revealed 5 different changes in NS1protein. Three of which are fixed amino acid changes E26K, D209N and K229E while 10 (31.3%) T58P and 5 (15.6%) A86S amino acid changes were also observed. There was no change observed in nuclear export protein (NEP). The study provides an insight on mutations in NS gene occurring in this viral gene in the country that has not been reported elsewhere.
Keywords: Influenza A (H3N2) virus, Iran, Mutation, NS1 gene.
Introduction:
Influenza viruses have been widely studied due to their pandemic capability. This capabilities have resulted in more mortality and severe disease in the general population, the most notable being the Spanish influenza pandemic of 1918 and 19191. Mechanism of virulence for these viruses is on their capacity to cause immunopathogenesis.
Synthesis of two mRNAs from the eighth vRNA segment of the viral gene was known by Influenza A virus. The early encodes NS1 protein, and the later is produced by splicing of NS1 mRNA that is translated into a protein which localizes in the cell nucleus that was formally named NS2 but were now renamed the nuclear export protein (NEP) 2,3.
NS1 protein is translated from the mRNA directly and consists of 124–237 amino acids (aa), depending on the virus strain 4- 6. The currently circulating Influenza A/H3N2 virus in Iranian population from this research is 230 aa-long. NS1 protein is made of two important domains: N-terminal RNA-binding domain (1–73) and C-terminal effector domain (73–237) residues 7. Though NS1 protein has different roles, one of its main actions is to alter type I IFN production by the host8, acts after transcription to prevent the 3´-end processing of host mRNA involving IFN mRNA by attaching to cleavage and polyadenylation specificity factor 30 (CPSF30) and poly-A-binding protein nuclear I 9,10 .
One hundred and twenty one amino acid-long proteins were translated from mRNA of NS2/NEP11. In the current study it is 120 aa-long. It may promote production of a stable export complex of new viral RNP. In alliance with matrix protein 1(M1), it interacts with cellular export factor (CEF1) and modulates nuclear export of viral ribonucleoprotein (vRNP) complexes by connecting cellular export machinery with vRNPs 14.
NS gene implication in virulence and replication of the virus is well-documented. It has been shown that both NS1 F103L and M106I were adaptive changes that promoted replicative capacities in cells of different species and also virulence in the mouse lung15. NS1 is one of multifunctional protein determinant of virulence with several functions in different ways to counteract the cellular innate immune response 14. It has been reported that H5N1 virus was able to interfere with IFN-β activation due to properties of its NS1 gene, inhabiting F103L and M106I changes and also nature of its NP and PA genes15. Avian influenza viruses sequence analysis shows that NS1 protein C-terminal four residues is a probable PDZ domain ligand (PL) of the X-S/T-X-V type 16.
Protein–protein recognition modulation that organizes diverse cell signaling assemblies was shown to be function PDZ domains. It was known to plays important roles in the cell recognition process. Ninety percent of the human influenza viruses with RSKV or RSEV in NS1 protein were shown to have a C-terminal four-residue PL sequence. High mortality outbreaks of the recent time known to be of avian origin and therefore contain avian-like NS1 C-terminal PL residues of ESEV or EPEV which are incriminated to about 7% of Homo sapiens viruses consist of H5N1 isolates. It was reported that avian viruses NS1 proteins bind to ≈30 known human PDZ domain-containing proteins, and NS1 proteins for human attaching to PDZ domain-containing proteins was not noticed 14. It has been reported that C terminus of the NS1 substitution with KSEV from the 1918 H1N1 virus increased pathogenicity as observed by morphological changes of lung sections, though the virus was shown to causes less weight loss in mice in contrast with viruses harboring the H5N1 HPAI14. Previous studies have identified a change in NS1 gene (S42P, D92E and V149A) with regard to the involvement of NS1 protein in virulence that increased viral pathogenicity17-19. NS gene extent of variation in the influenza A (H3N2) viruses in Iran in recent years has not been described. Presently analysis of NS gene of 32 isolates where carried out from Iranian population in order to secure more detailed information about its phylogeny and genetic changes, and compare the changes found in these strains, the vaccine strain and other isolates from countries in northern hemisphere. The aim of the study is sequence analysis of amino acid and identification of changes in the NS gene of influenza A (H3N2) virus isolated in Iran that may affect the functions of this protein in pathogenesis.
Methods:
Two hundred and fourteen specimens of patients with respiratory illness were obtained from the National Influenza Centre at School of Public Health, Tehran University of Medical Sciences. It was cultured in MDCK cell line following WHO guide line for influenza A virus isolation. RNA extraction was carried out with High Pure Viral Nucleic acid extraction kit (Roche Diagnostic, Germany) according to manufacturer’s protocol. Real-Time RT-PCR was carried out for the detection of Influenza viruses. Ninety specimens were positive for influenza A/H3N2 virus.
NS gene was amplified using one-step RT-PCR kit (Qiagen) and specific primers (NSH3N2-F:5´-AGCAAAAGCAGGGTGACAAAGA-3´ and NSH3N2-R:5´-GAGAAAGTTCTTATCTCCTGTTCCA-3´). The condition for the RT-PCR was as follows: Reverse transcription at 50°C for 40 minutes, initial PCR activation at 95°C for 10 minutes, Denaturation at 95°C for 30 second, Annealing at 57°C for 30 second, Extension at 72°C for 1 minutes (for 40 cycles) and a final extension at 75°C for 10 minutes. The PCR products were purified using Qiaquick gel purification kit (Qiagen) according to manufacturer’s instruction. Samples were sequenced using specific primers and Big Dye terminator v3.1 cycle sequencing kit with the genetic analyzer ABI 3130 in the National Influenza Laboratory. Sequence alignment was done using BIOEDIT and ClustalW program20. Sequence alignment revealed similarities amongst the isolates. NS gene sequences of 32 influenza A/H3N2 virus strains isolated in this study out of 90 sequenced were selected base on the site of sample collection, compared and analyzed with the vaccine strain and the sequences obtainable from other countries in the northern hemisphere already deposited in GenBank. Phylogenetic tree construction were performed using MEGA Software version 5.05 21.
Result
Isolates represent samples collected from June 2013 to March 2014. In the current study 230 aa were found in NS1 while the NS2/NEP had 120 amino acids. Analysis depicted relationship phylogenetically that shows there are exact clusters of the virus suggestive of co-circulation of multiple sub-lineages (Figure 1).
In respect to genetic variation, H3 numbering system of amino acid was used in naming the detected amino acid changes 13, 14. Substitutions were found in 26, 58, 86, 209 and 229 aa positions in NS1 protein, respectively (Table 1). There was no amino substitution in regard to NS2/NEP found. NS1 proteins have fixed amino acid substitution at positions 26, 209 and 229 in the current study. At position 26, it occurs as a result of substitution at the 76-78 Codon from GAA to AAA causing a glutamate to lysine substitution. In position 209, substitution occurs at 625-627 Codon from GAT to AAT causing an aspartate to asparagine substitution. While in position 229, substitution occurs at 685 – 687 Codon from AAA to GAA causing a lysine to glutamate substitution. Another 10 (30.3%) NS1 proteins substitution occurs at (58 amino acid position) 172-174 Codon from ACC to CCC causing a threonine to proline substitution. Also 5 (15.2%) of the NS1 proteins substitution occurs at (amino acid position 86) 256-258 Codon from GCT to TCT causing an alanine to serine substitution.
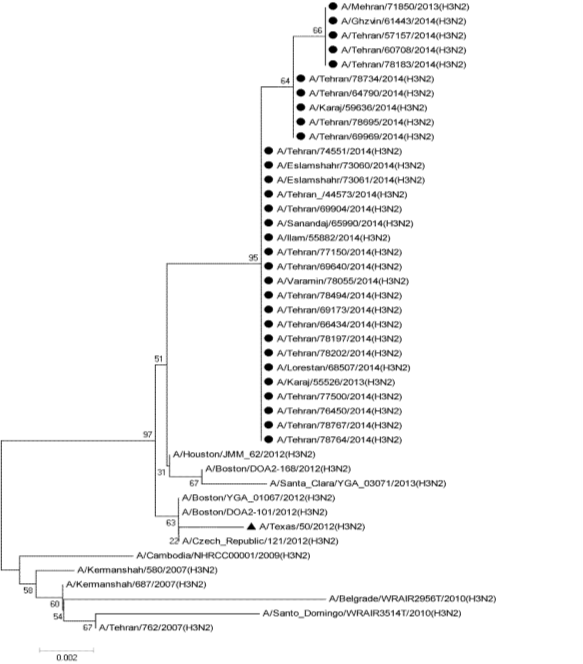
Figure 1 Relationship of the NS nucleotide coding region phylogenetically of influenza viruses used in this study. Neighbor-joining analysis with Tamura-Neibour model, using MEGA 5.05 was used to generate the tree. Circular black dot represent studied samples and triangular black dot represent vaccine strain.
Table 1: Amino acid changes of NS1 protein of influenza A/H3N2 virus strains from Iranian Population compared with vaccine strain and other countries in northern hemisphere.
Virus isolates Amino acid at indicated position in NS1 protein
1 1 1 1 1 1 2 2 2
2 5 8 8 8 1 2 3 3 4 6 0 0 2
6 8 0 5 6 1 9 5 9 0 4 4 9 9
|
A/Texas/50/2012 |
E |
T |
T |
F |
A |
V |
M |
N |
G |
R |
P |
R |
D |
K |
|
A/Tehran/77254/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Eslamshahr/73061/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Eslamshahr/73060/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/74551/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran /44573/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/69904/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Sanandaj/65990/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Ilam/55882/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/77150/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran /78183/2014 |
K |
P |
• |
• |
S |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/60708/2014 |
K |
P |
• |
• |
S |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/57157/2014 |
K |
P |
• |
• |
S |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/69969/2014 |
K |
P |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/69640/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Varamin/78055/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/78494/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/69173/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/66434/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/78197/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/78202/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Lorestan/68507/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Karaj 2/55526/2013 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Mehran/71850/2013 |
K |
P |
• |
• |
S |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Ghazvin/61443/2014 |
K |
P |
• |
• |
S |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/78695/2014 |
K |
P |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/77500/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Karaj/59636/2014 |
K |
P |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/76450/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/64790/2014 |
K |
P |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/78734/2014 |
K |
P |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/78767/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/78764/2014 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Belgrade/WRAIR2956T/2010 |
• |
• |
A |
L |
• |
• |
• |
S |
D |
• |
H |
• |
N |
• |
|
A/S.Domingo/WRAIR3514T/2010 |
• |
• |
• |
• |
• |
• |
T |
S |
D |
Q |
• |
K |
N |
• |
|
A/Houston/JMM_62/2012 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Boston/YGA_01067/2012 |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Tehran/762/2007(H3N2) |
• |
• |
• |
• |
• |
• |
• |
S |
D |
• |
• |
• |
N |
• |
|
A/Kermanshah/687/2007(H3N2) |
• |
• |
• |
• |
• |
• |
• |
S |
D |
• |
• |
• |
N |
• |
|
A/Kermanshah/580/2007(H3N2) |
• |
• |
• |
• |
• |
• |
• |
S |
D |
• |
• |
• |
N |
• |
|
A/Boston/DOA2-101/2012 |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Czech Republic/121/2012 |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Boston/DOA2-168/2012 |
K |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Cambodia/NHRCC00001/2009 |
K |
• |
• |
• |
• |
M |
• |
S |
D |
• |
• |
• |
N |
• |
|
A/Santa Clara/YGA_03071/2013 |
K |
• |
• |
• |
• |
L |
• |
• |
• |
• |
• |
• |
N |
E |
|
A/Boston/YGA_01067/2012 |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
• |
N |
E |
NS2/NEP analysis does not reveal any amino acid change in the present study (Table 2).
Nucleotide sequence data obtained has been deposited to GenBank database and can be retrieved under accession numbers KP162026- KP162056 and KP209320.
Table 2: Amino acid changes of NS2/NEP protein of influenza A/H3N2 virus strains from Iranian Population compared with vaccine strain and other countries in northern hemisphere.
|
Virus isolates |
Amino acid at indicated |
position in NS2 protein |
|
47 |
88 |
|
|
A/Texas/50/2012 |
E |
R |
|
A/Tehran/77254/2014 |
• |
• |
|
A/Eslamshahr/73061/2014 |
• |
• |
|
A/Eslamshahr/73060/2014 |
• |
• |
|
A/Tehran/74551/2014 |
• |
• |
|
A/Tehran /44573/2014 |
• |
• |
|
A/Tehran/69904/2014 |
• |
• |
|
A/Sanandaj/65990/2014 |
• |
• |
|
A/Ilam/55882/2014 |
• |
• |
|
A/Tehran/77150/2014 |
• |
• |
|
A/Tehran /78183/2014 |
• |
• |
|
A/Tehran/60708/2014 |
• |
• |
|
A/Tehran/57157/2014 |
• |
• |
|
A/Tehran/69969/2014 |
• |
• |
|
A/Tehran/69640/2014 |
• |
• |
|
A/Varamin/78055/2014 |
• |
• |
|
|
You have to be 100% sure of the quality of your product to give a money-back guarantee. This describes us perfectly. Make sure that this guarantee is totally transparent.
Read moreEach paper is composed from scratch, according to your instructions. It is then checked by our plagiarism-detection software. There is no gap where plagiarism could squeeze in.
Read moreThanks to our free revisions, there is no way for you to be unsatisfied. We will work on your paper until you are completely happy with the result.
Read moreYour email is safe, as we store it according to international data protection rules. Your bank details are secure, as we use only reliable payment systems.
Read moreBy sending us your money, you buy the service we provide. Check out our terms and conditions if you prefer business talks to be laid out in official language.
Read more